pacman::p_load(psych, GPArotation, tidyverse, readxl, gt, gtExtras, dendextend, cluster, factoextra, useful, ggrepel)
# コード11-1
# install.packages("psych")
# install.packages("GPArotation")
# コード11-2
factor_exdata <- readxl::read_excel("data/factor_ex.xlsx", na = ".")
factor_exdata$V5 <- 8 - factor_exdata$V5
rownames(factor_exdata) <- factor_exdata$ID
factor_exdata2 <- factor_exdata %>%
select(-ID)
knitr::kable(summary(factor_exdata2))| V1 | V2 | V3 | V4 | V5 | V6 | |
|---|---|---|---|---|---|---|
| Min. :1.000 | Min. :2.0 | Min. :1.0 | Min. :2.0 | Min. :1.0 | Min. :2.000 | |
| 1st Qu.:2.000 | 1st Qu.:3.0 | 1st Qu.:2.0 | 1st Qu.:3.0 | 1st Qu.:3.0 | 1st Qu.:3.000 | |
| Median :4.000 | Median :4.0 | Median :4.0 | Median :4.0 | Median :4.5 | Median :4.000 | |
| Mean :3.933 | Mean :3.9 | Mean :4.1 | Mean :4.1 | Mean :4.5 | Mean :4.167 | |
| 3rd Qu.:6.000 | 3rd Qu.:5.0 | 3rd Qu.:6.0 | 3rd Qu.:5.0 | 3rd Qu.:6.0 | 3rd Qu.:4.750 | |
| Max. :7.000 | Max. :7.0 | Max. :7.0 | Max. :7.0 | Max. :7.0 | Max. :7.000 |
| V1 | V2 | V3 | V4 | V5 | V6 | |
|---|---|---|---|---|---|---|
| V1 | 1.0000000 | -0.0532178 | 0.8730902 | -0.0861622 | 0.8576366 | 0.0041681 |
| V2 | -0.0532178 | 1.0000000 | -0.1550200 | 0.5722121 | -0.0197456 | 0.6404649 |
| V3 | 0.8730902 | -0.1550200 | 1.0000000 | -0.2477879 | 0.7778480 | -0.0180688 |
| V4 | -0.0861622 | 0.5722121 | -0.2477879 | 1.0000000 | 0.0065819 | 0.6404649 |
| V5 | 0.8576366 | -0.0197456 | 0.7778480 | 0.0065819 | 1.0000000 | 0.1364029 |
| V6 | 0.0041681 | 0.6404649 | -0.0180688 | 0.6404649 | 0.1364029 | 1.0000000 |
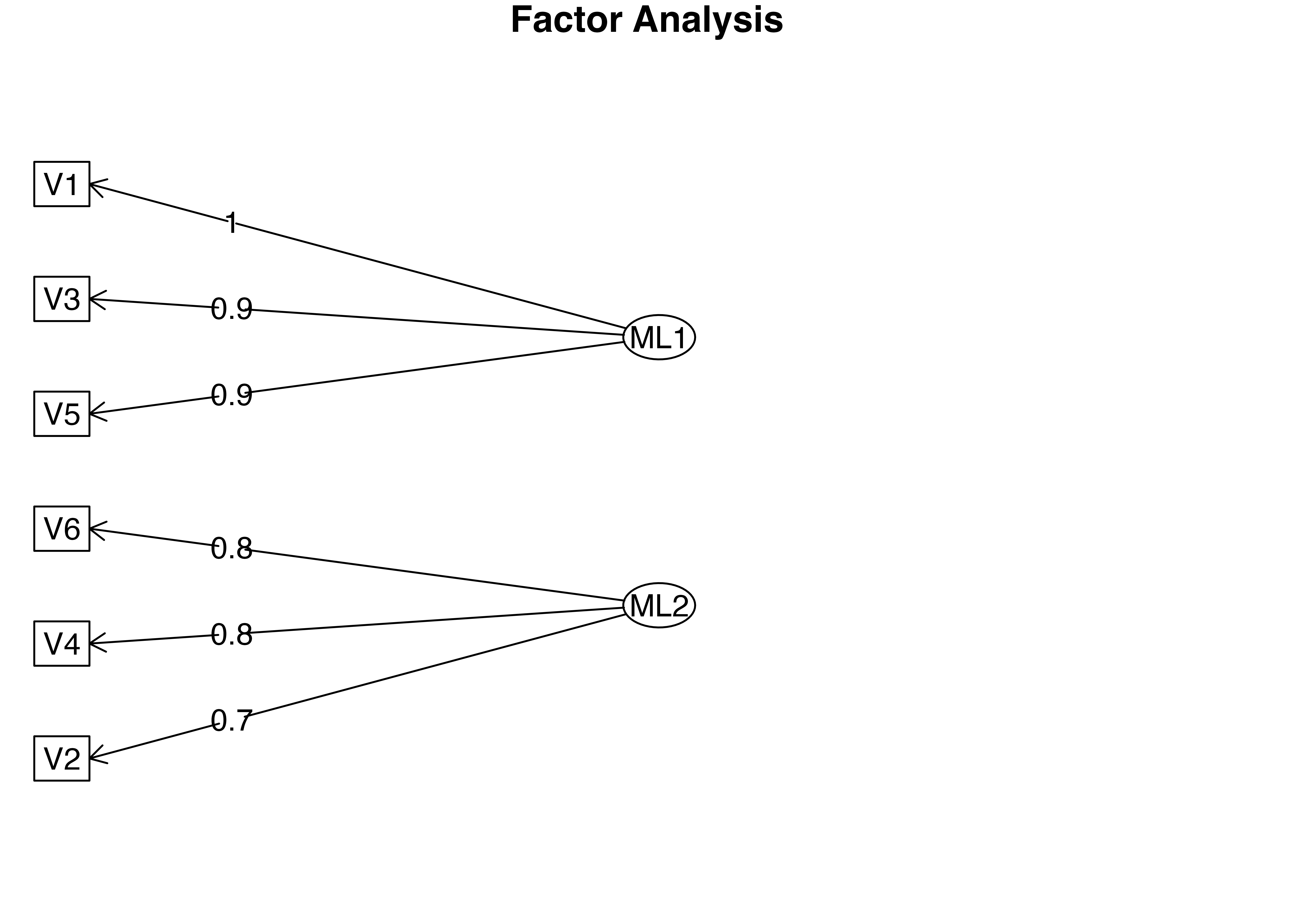
Factor Analysis using method = ml
Call: fa(r = factor_exdata2, nfactors = 2, rotate = "promax", fm = "ml")
Standardized loadings (pattern matrix) based upon correlation matrix
ML1 ML2 h2 u2 com
V1 0.97 0.00 0.94 0.063 1
V2 -0.02 0.75 0.56 0.437 1
V3 0.89 -0.12 0.83 0.174 1
V4 -0.06 0.78 0.62 0.378 1
V5 0.89 0.11 0.79 0.205 1
V6 0.08 0.83 0.69 0.309 1
ML1 ML2
SS loadings 2.54 1.89
Proportion Var 0.42 0.32
Cumulative Var 0.42 0.74
Proportion Explained 0.57 0.43
Cumulative Proportion 0.57 1.00
With factor correlations of
ML1 ML2
ML1 1.00 -0.06
ML2 -0.06 1.00
Mean item complexity = 1
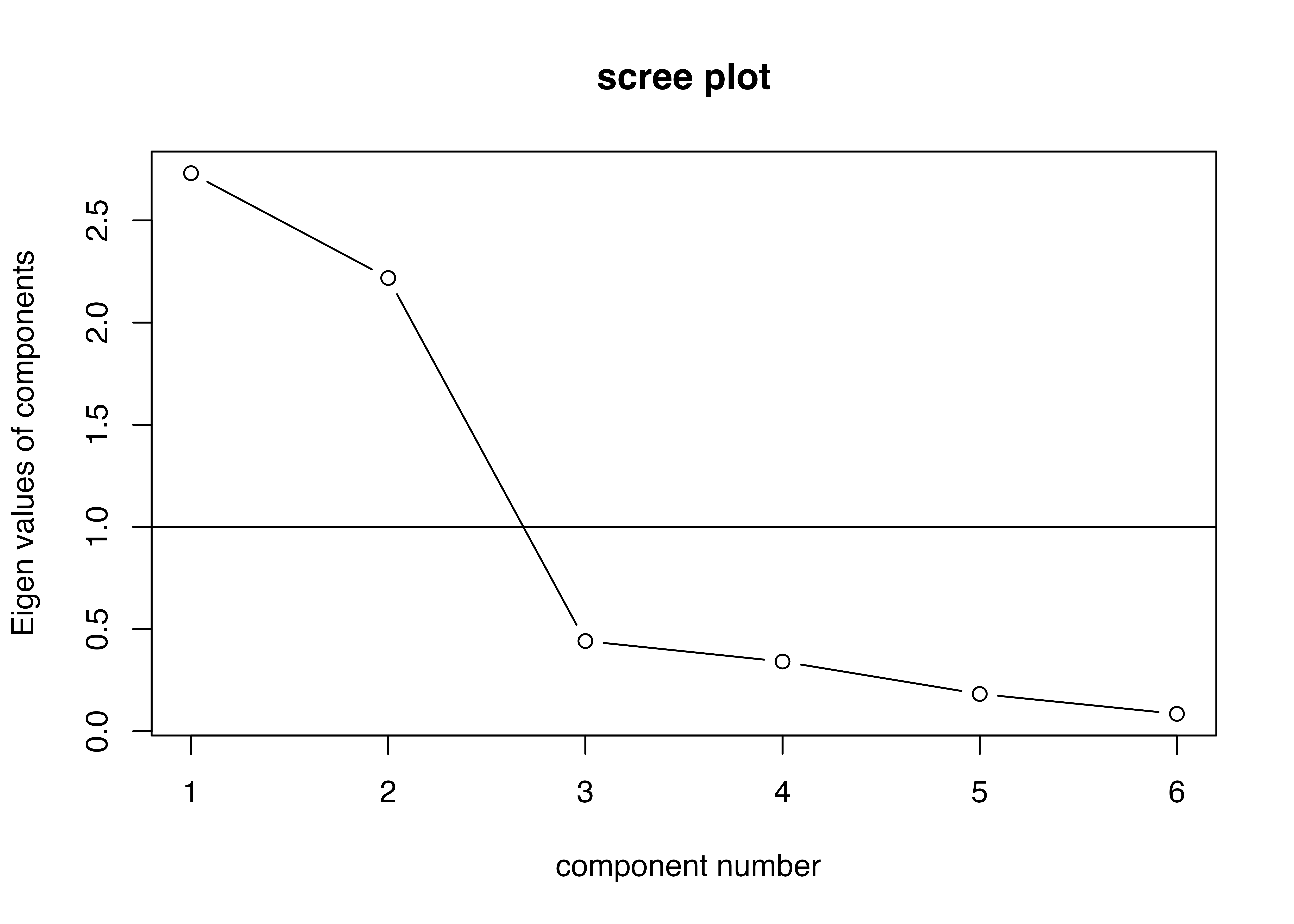
Test of the hypothesis that 2 factors are sufficient.
df null model = 15 with the objective function = 4.25 with Chi Square = 111.31
df of the model are 4 and the objective function was 0.21
The root mean square of the residuals (RMSR) is 0.03
The df corrected root mean square of the residuals is 0.05
The harmonic n.obs is 30 with the empirical chi square 0.65 with prob < 0.96
The total n.obs was 30 with Likelihood Chi Square = 5.21 with prob < 0.27
Tucker Lewis Index of factoring reliability = 0.95
RMSEA index = 0.095 and the 90 % confidence intervals are 0 0.314
BIC = -8.39
Fit based upon off diagonal values = 1
Measures of factor score adequacy
ML1 ML2
Correlation of (regression) scores with factors 0.98 0.92
Multiple R square of scores with factors 0.96 0.84
Minimum correlation of possible factor scores 0.92 0.68| V1 | V2 | V3 | V4 | V5 | V6 | rowname | ML1 | ML2 |
|---|---|---|---|---|---|---|---|---|
| 7 | 3 | 6 | 4 | 6 | 4 | 1 | 1.3106155 | -0.2896470 |
| 1 | 3 | 2 | 4 | 3 | 4 | 2 | -1.2878038 | -0.2073067 |
| 6 | 2 | 7 | 4 | 7 | 3 | 3 | 1.1828544 | -0.7996332 |
| 4 | 5 | 4 | 6 | 6 | 5 | 4 | 0.1474225 | 1.0035837 |
| 1 | 2 | 2 | 3 | 2 | 2 | 5 | -1.3907544 | -1.3067260 |
| 6 | 3 | 6 | 4 | 6 | 4 | 6 | 0.9928111 | -0.2875982 |
# コード11-9
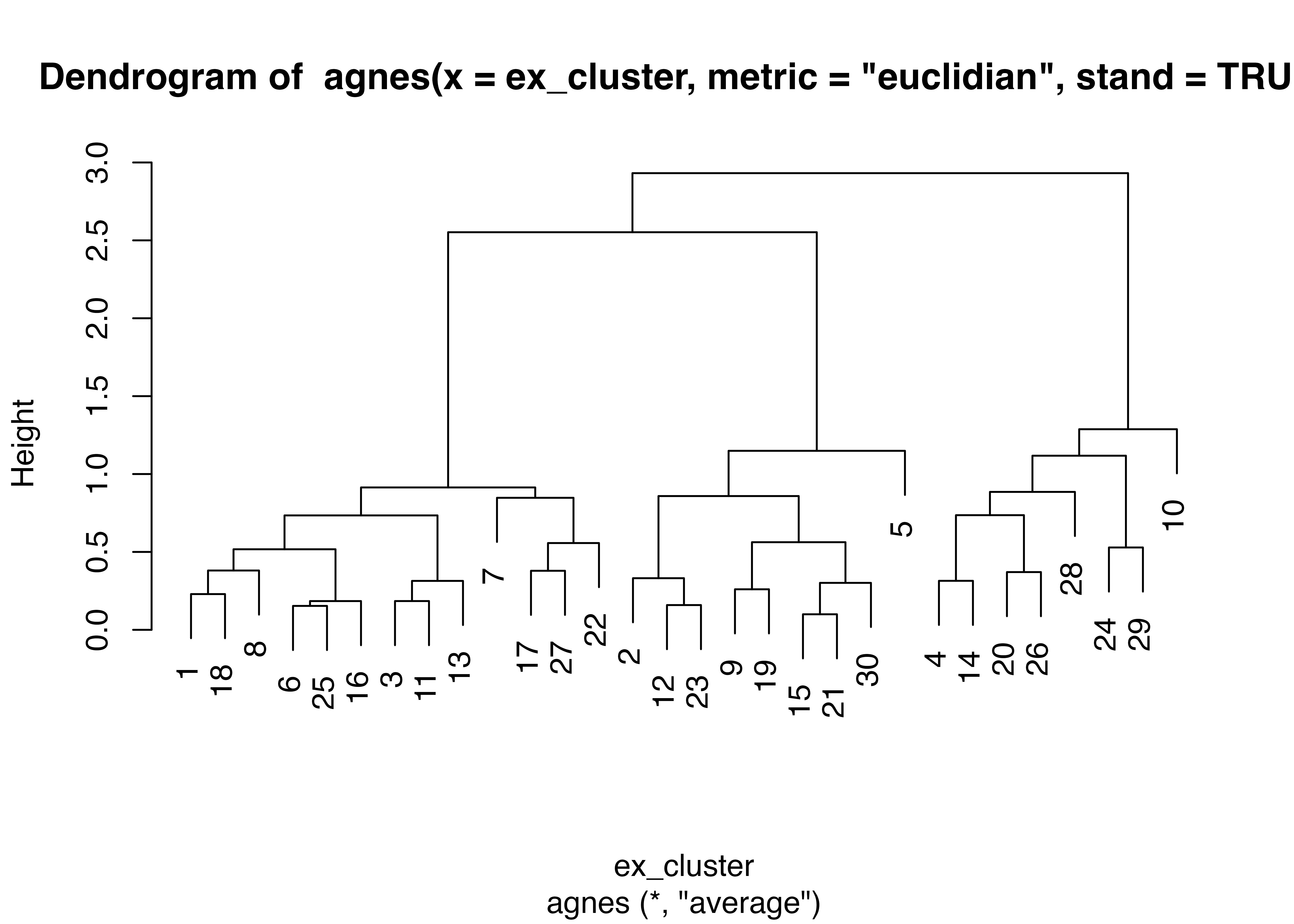
cl_1 <- kmeans(ex_cluster, 3)
clus_fa <- data.frame(cl_1$cluster)
clus_fa$rowname <- rownames(clus_fa)
factor_exdata2 <- left_join(factor_exdata2, clus_fa, by = "rowname")
factor_exdata2$cl_1.cluster <- factor(factor_exdata2$cl_1.cluster)
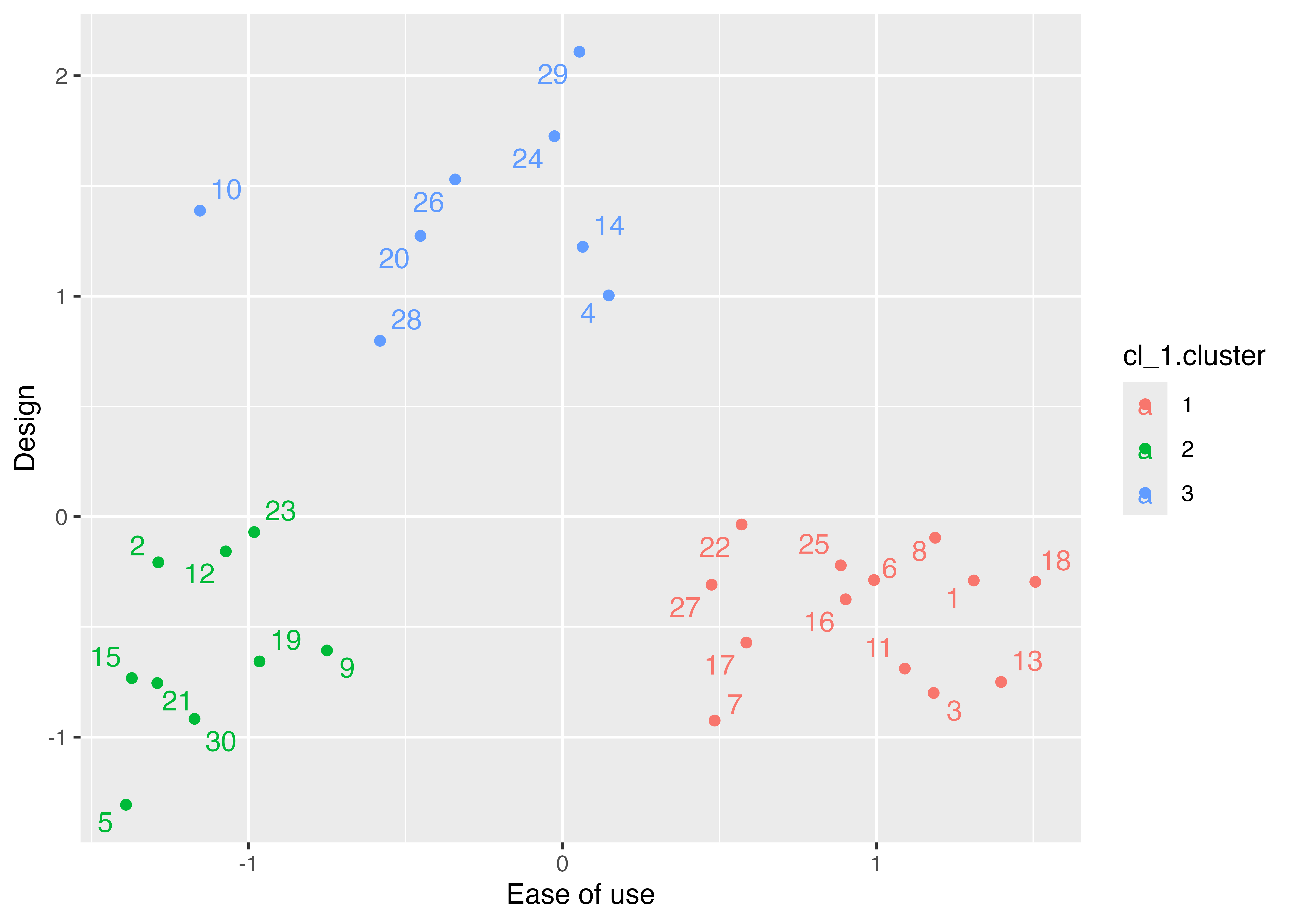
# Visualizing the clusters with 2 factors
p1 <- ggplot(data = factor_exdata2,
mapping = aes(x = ML1, y = ML2, color = cl_1.cluster))
p1 + geom_point() +
geom_text_repel(mapping = aes(label = rownames(factor_exdata2))) +
labs(x = "Ease of use", y = "Design")Principal Components Analysis
Call: principal(r = r, nfactors = nfactors, residuals = residuals,
rotate = rotate, n.obs = n.obs, covar = covar, scores = scores,
missing = missing, impute = impute, oblique.scores = oblique.scores,
method = method, use = use, cor = cor, correct = 0.5, weight = NULL)
Standardized loadings (pattern matrix) based upon correlation matrix
PC1 PC2 h2 u2 com
V1 0.93 0.25 0.93 0.074 1.1
V2 -0.30 0.80 0.72 0.277 1.3
V3 0.94 0.13 0.89 0.106 1.0
V4 -0.34 0.79 0.74 0.261 1.4
V5 0.87 0.35 0.88 0.122 1.3
V6 -0.18 0.87 0.79 0.210 1.1
PC1 PC2
SS loadings 2.73 2.22
Proportion Var 0.46 0.37
Cumulative Var 0.46 0.82
Proportion Explained 0.55 0.45
Cumulative Proportion 0.55 1.00
Mean item complexity = 1.2
Test of the hypothesis that 2 components are sufficient.
The root mean square of the residuals (RMSR) is 0.07
with the empirical chi square 3.94 with prob < 0.41
Fit based upon off diagonal values = 0.98# コード11-11
# データの相関行列から固有値と固有ベクトルを抽出
cor_ex <- cor(factor_exdata3)
eigen_list <- eigen(cor_ex)
# 固有ベクトルについての情報をオブジェクトとして定義
pc1_eig <- eigen_list$vectors[, 1]
pc2_eig <- eigen_list$vectors[, 2]
# 固有ベクトルに,固有値の平方根を乗じる
PC1 <- pc1_eig * sqrt(eigen_list$values[1])
PC2 <- pc2_eig * sqrt(eigen_list$values[2])
knitr::kable(data.frame(cbind(PC1, PC2)), caption = "計算結果")| PC1 | PC2 |
|---|---|
| 0.9283425 | -0.2532285 |
| -0.3005297 | -0.7952496 |
| 0.9361812 | -0.1308894 |
| -0.3415817 | -0.7889663 |
| 0.8687553 | -0.3507939 |
| -0.1766389 | -0.8711581 |